Study Uncovers a Strategy that Viruses use to Evade Immediate Immune Responses
PR Newswire
WASHINGTON, Aug. 23, 2019
WASHINGTON, Aug. 23, 2019 /PRNewswire/ — Researchers at the U.S. Naval Research Laboratory have identified a new algorithm that could help virologists better understand and treat infections caused by viruses like the Zika virus.
Patricia Legler, a research biologist at NRL, discovered the algorithm while developing Venezuelan Equine Encephalitis virus (VEEV) protease assays for drug discovery. One of the substrates she designed contained a human protein sequence that she thought could be cut by the viral protease.
After searching for similar cases, Legler found that most of the viral proteases that could cut host proteins were from positive sense single-stranded RNA viruses. She also noticed that the proteins that were being cut were involved in generating the innate, or immediate, immune responses to these viruses. Legler’s findings showed that these viruses used what she called a “search and delete” algorithm to target and destroy host proteins.
“The virus’s protease finds the key word—or protein sequence—[in a host cell] and then deletes the file containing the keyword, this is one mechanism that allows the virus to replicate,” Legler said.
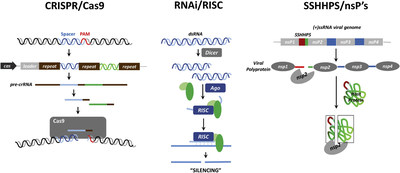
While studying how these viruses replicate, Legler realized that the procedures her team created to emulate a virus had uncovered an efficient way to describe how a virus edits a host’s proteome. The process closely resembles that used by the CRISPR-Cas9 enzyme to destroy invading strands of phage DNA.
Also contributing to this study were Xin Hu and Juan Marugan of National Center for Advancing Translational Sciences (NCATS/NIH), and Elaine Morazzani and Pamela Glass from U.S. Army Medical Research Institute of Infectious Diseases (USAMRIID).
Their research findings were published in an issue of Elsevier’s Antiviral Research in April (DOI: https://doi.org/10.1016/j.antiviral.2019.02.001).
Finding the targets of a virus
Using established cultural cell lines, Legler’s study identified human protein sequences that could be “cut” by the protease of the Zika virus. The “keywords” are what researchers call cleavage sites— in this case, the keywords were in proteins related to immunity. The viruses “delete” these proteins, allowing the virus to replicate and spread.
“That means that these aren’t just any proteins being cut,” Legler said. “This is a targeted strategy to antagonize your innate immune responses. That leads us to the idea that this cleavage site sequence actually corresponds to a host protein sequence.”
Legler labeled these protein sequences short stretches of homologous host-pathogen protein sequences (SSHHPS) to acknowledge NRL. Some of the protein sequences that could be cut by the Zika protease were in proteins called FOXG1, NT5M and SFRP1. These proteins are involved in brain and eye development. FOXG1 mutations have been associated with a syndrome characterized by microcephaly, an unusually small head size.
The algorithm the researchers uncovered identified new protein sequences that could aid biologists in understanding human antiviral defense systems and help determine which pathways the virus targets. “We think we can actually use this to predict what the virus will do to the host,” Legler said.
A new way to create antivirals
Researchers can also use the algorithm to identify animal models by matching the proteins targeted by the virus in the human body with those in animals. Legler discovered species-specific differences in the cleavage sites while studying the Venezuelan equine encephalitis virus in tandem with Zika.
The Venezuelan equine encephalitis virus, or VEEV, is aggressive in killing horses—with a 20-80 percent mortality rate. It is less successful in killing humans who have only a 1 percent mortality rate. Using the algorithm to study the virus, Legler learned that horses lack part of an innate immune response protein that humans have that helps the human body fight VEEV.
“That indicates that species with a cleavage site that matches to the virus have some kind of defense mechanism for these infections,” she explained. “Whereas the species that don’t have the cleavage site may be the most susceptible to this disease.”
The sequence differences may also make other species less likely to develop certain viral infections.
Legler’s algorithm will allow researchers to compare the protein sequences attacked by a virus in the human body with databases of animal proteins to find matching sequences. The hope is that matching the protein sequences of animal models with those in humans could allow virologists to test antiviral medications and vaccines that are more effective at combating the effects of a virus.
Moving the algorithm forward
While the algorithm provided Legler with valuable information about the viruses she studied and a way to replicate their “search and delete” programs, she and her collaborators did much of their work by hand.
“When I was studying the Zika virus, I compiled a table of all of the host proteins that have been shown to be cut and drew them out,” she said.
Legler and her collaborators hope to streamline this process by creating software to carry out the algorithm, removing the need to make the tables by hand.
“By creating software, we can find a way to provide other scientists with an easy interface to identify the host protein sequences in the viruses they’re studying and parallel animal models,” she explained. “[That will give] them a functional tool to use as they create antivirals.”
The U.S. Naval Research Laboratory provides the advanced scientific capabilities required to bolster our country’s position of global naval leadership. With more than 2,500 personnel scientists, engineers and support staff, it has served the U.S. Navy and the nation for nearly 100 years, advancing research further than you can imagine. For more information, visit the NRL website and join the conversation on Twitter, Facebook, and YouTube.
By Gabrielle M. Gibert | Strategic Communications, U.S. Naval Research Laboratory
Editor Contact: Daniel Parry, U.S. Naval Research Laboratory Public Affairs
[email protected]
202-767-2326

![]() View original content to download multimedia:http://www.prnewswire.com/news-releases/study-uncovers-a-strategy-that-viruses-use-to-evade-immediate-immune-responses-300906285.html
View original content to download multimedia:http://www.prnewswire.com/news-releases/study-uncovers-a-strategy-that-viruses-use-to-evade-immediate-immune-responses-300906285.html
SOURCE U.S. Naval Research Laboratory
